Class Meeting 17 (3) Functional programming in R: Part I
17.1 Today’s Agenda
- Announcements:
- Reminder about Assignment 1 and 2
- Milestone 2 is now posted
- Office hours are on Tuesday & Thursday after class in this room, and on Fridays from 10-12 in ESB 1045.
- Download today’s participation file (and commit into your participation repo)
- Part 1: Introduction to functional programming (FP) (10 mins)
- Motivation for functions and for vectorizing operations
- Anatomy of a function in R
- Comments on RScript vs. RMarkdown vs. RNotebook
- Part 2: Vectorization
- What is vectorization?
- Why do we use vectorization?
- Examples of vectorized operations in R
- Part 3: Functional programming using the
purrrpackagepurrr::map- Use the right
purrr::map*function based on your desired output - Specify some arguments of the function
- Mapping with two data objects
- Mapping with more than two data objects
17.2 Learning outcomes for this lecture
- Define the philosophy of functional programming in R.
- Describe the benefits of vectorizing R code.
- Apply vectorization to tasks in R.
- List and describe the
mapfunctions from thepurrrpackage. - Apply functions from the
purrrpackage to vectorize tasks in R. - Describe anonymous functions, apply them, and use the shorthand notation in
purrrfunctions.
17.3 Part 1: Introduction to functional programming (FP) (10 mins)
17.3.1 Motivation for functions and for vectorizing operations
17.3.2 Anatomy of a function in R

func_name <- function (arg1, arg2 = 5, arg3 = TRUE) {
if (arg3 == TRUE) {
print(glue::glue(arg1," will be raised to the power of ", arg2))
}
arg1^arg2
}
func_name(arg1 = 4, arg2 = 3, arg3 = FALSE)## [1] 64## Error in glue::glue(arg1, " will be raised to the power of ", arg2): argument "arg1" is missing, with no default17.3.3 Rscript (.R) vs. RMarkdown (.Rmd) vs. RNotebook (Rmd + special YAML Header)
This site has some nice visuals that show you differences between an Rmarkdown document and an RNotebook. Rscripts are not interactive and designed to be run from the command line.
17.4 Part 2: Vectorization
Many thanks to one of our teaching assistants Sirine Chahma for the first draft of this lecture!
17.4.1 What is vectorization?
There are several ways of applying the same operation to all the elements of a given vector.
You can “brute force” it:
## [1] 2 4 6 8But it’s very easy to make mistakes when you’re copy/pasting code like this so it’s a good rule of thumb to think of better ways to do things when you have to copy and paste the same block of code more than about once.
Let’s try this again:
## [1] 2 4 6 8Rats! We made another mistake. Find and fix the mistake in the code above please!
Okay, let’s get to the better way of doing things.
You can use a loop :
## [1] 1 2 3 4## [1] 2 4 6 8There is a function called seq_along that essentially replaces 1:length(x) in the code chunk above:
## [1] 2 4 6 8We will use seq_along(x) and 1:length(x) interchangeably.
So, this works and is much less error-prone, but in this case - there is actually an even better option, - vectorized operations! Let’s see an example of it:
## [1] 2 4 6 8You might have thought this was an obvious thing to try, and you’d be right - R has some built in functions to handle vectorization “behind the scenes”. For example, we can sum the values of two vectors :
x1 <- c(1, 2, 3, 4)
x2 <- c(10, 20, 30, 40)
y <- c()
for (i in 1:length(x1)){
y[i] <- x1[i] + x2[i]
}
y## [1] 11 22 33 44but built-in vectorization in R allows us to do this:
## [1] 11 22 33 4417.4.2 Why do we use vectorization?
Let’s come back to the first example we saw (multiply the values of a vector by 2), but let’s use a bigger vector this time.
## The length of x is 100000000Take a guess at how long the loop below is going to take to run (Hint: the answer is “in the seconds”)?
# Guess at how long this loop takes
x <- 1:100000000
for (i in 1:length(x)){
y[i] <- 2*x[i]
}
## YOUR GUESS HERELet’s try using the tictoc package to time how long this operation takes.
tic starts the clock, and toc stops the clock and prints out the total time.
Let’s take a look at the time taken by the vectorized operation now :
## 1.37 sec elapsedWow! That is amazing - see how much faster the vectorized operation is compared to the for loop.
It’s usually recommended to use vectorized operation rather than regular loops for several reasons, including memory efficiency, speed, readability, “debugability”, and easily being able to add tests (more on this next week).
17.4.3 Examples of vectorized operations
Here are a few examples of other operations that are vectorized.
- Check if the values of two vectors are the same :
x1 <- c(1, 2, 3, 4)
x2 <- c(1, 2, 1, 2)
y <- x1 == x2
# Can you guess the values of `y`?
print(c(TRUE, TRUE, FALSE, FALSE))## [1] TRUE TRUE FALSE FALSEAnd the answer is (run in RStudio):
- Compare the values of two vectors :
x1 <- c(1, 2, 3, 4)
x2 <- c(1, 2, 1, 2)
y <- x1 > x2
# Can you guess the values of `y`?
print("YOUR SOLUTION HERE")## [1] "YOUR SOLUTION HERE"And the answer is:
- Logical comparaisons can also be used:
# compares each elements of each vector by position
y <- c(TRUE, TRUE, TRUE) & c(FALSE, TRUE, TRUE)
y## [1] FALSE TRUE TRUEThere are a lot of other operations that are vectorized! Here is a list of vector operators : R Operators cheat sheet
17.5 Part 3: Functional programming using the purrr package
Until now, we have just applied simple operations to vectors. The functions were only applied to a single element of the vector, which were doubles. What if we want to use data frames (as you likely will in your projects)? In this case, one “element” becomes a whole vector (a column of the data frame), and the functions have to accept a vector as an input.
Let’s now try to work with data frames. How do we apply a function to all the columns of a data frame?
We are going to work with the iris data frame :
#select only the columns that represents a numerical variable
iris_df <- iris %>%
select(-Species)
head(iris_df)## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 5.1 3.5 1.4 0.2
## 2 4.9 3.0 1.4 0.2
## 3 4.7 3.2 1.3 0.2
## 4 4.6 3.1 1.5 0.2
## 5 5.0 3.6 1.4 0.2
## 6 5.4 3.9 1.7 0.4Let’s compute the mean of each column using a for loop :
means <- vector("double", ncol(iris_df))
## YOUR SOLUTION HERE
for (i in seq_along(iris_df)) {
means[i] <- mean(iris_df[[i]], na.rm = TRUE)
}means contains the means of each column :
We can do the same to find the minimum of each column :
mins <- vector("double", ncol(iris_df))
## YOUR SOLUTION HERE
for (i in seq_along(iris_df)) {
mins[i] <- min(iris_df[[i]], na.rm = TRUE)
}mins contains the minimums of each column :
The two loops we just wrote seem to very similar to each other, we should try to write a function that takes the function we want to apply and a data frame as its inputs.
my_function <- function(x, fun) {
value <- vector("double", ncol(x))
for (i in seq_along(x)) {
value[i] <- fun(x[[i]], na.rm = TRUE)
}
value
}Let’s check if we find the same values as before.
Try calling my_function to compute the mean and min of iris_df:
## [1] 5.843333 3.057333 3.758000 1.199333## [1] 4.3 2.0 1.0 0.1We find exactly the same values as when we were using the for loop!
Note: We have just written a functional, which is a function that takes another function as an input, and returns a vector as an output.
Just as a preview, the purrr package has some really convenient function(al)s that allow us to pass in other functions to apply to data frame.
17.5.1 The most general purrr function: map
The purrr:map function takes at least two arguments : a data frame and a function.
map(.x, .f, ...)
This means that we are going to apply the function f for every element of x.
This image may help you to better understand what does the purrr:map function does :
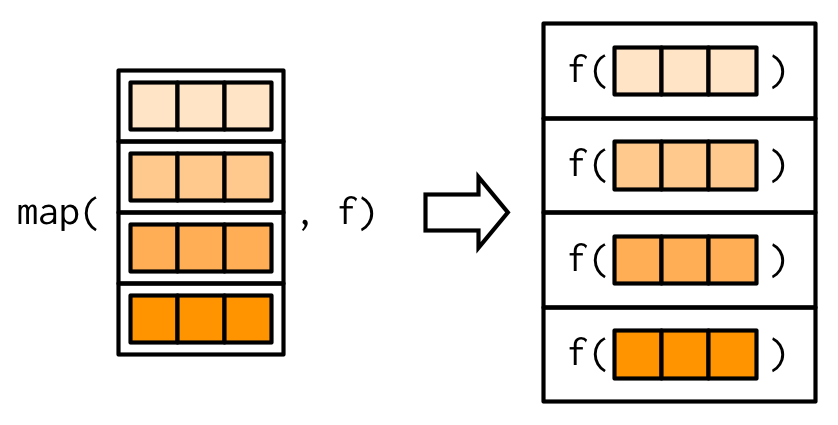 Source: Advanced R by Hadley Wickham.
Source: Advanced R by Hadley Wickham.
Note : In this image, the elements of the object that are used as an input seem to be the rows, but when we use a data frame as the input, they actually correspond to the columns of the data frame.
Let’s calculate the mean of the columns of the iris data frame :
## $Sepal.Length
## [1] 5.843333
##
## $Sepal.Width
## [1] 3.057333
##
## $Petal.Length
## [1] 3.758
##
## $Petal.Width
## [1] 1.199333The only difference with our my_function function we created above is that the output is a list!
17.5.2 Use the right purrr::map* function based on your desired output
Now, let’s take a look at the other functions that exist in the purrr library.
Here is a cheatsheet that contains a list of all the functions, and how to use them.
We have map_chr (character vector), map_dbl (double/numeric vector), map_dfc (dfc for dataframe columns and dfr for dataframe rows), map_int (integer) and map_lgl (logical).
Let’s practice with purrr:map_dbl:
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 5.843333 3.057333 3.758000 1.199333This time, the output is a vector containing doubles! This is exactly what we had with the function we created.
What if we want to specify some arguments of our function (ignore the NAs when we compute the mean for instance)?
We need to do a bit of work to do that - essentially we need to tell the map functional to also consider the na.rm argument of the mean function.
Let’s see how…
17.5.3 Specify some arguments of the function
Let’s introduce some missing data in our data frame :
What happens if we use purrr:map_dbl?
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## NA 3.057333 3.758000 1.199333The mean of the first column is now equal to NA. To solve this issue, we can use na.rm = TRUE as an argument of the mean function. But how do we add this to our map_dbl call?
We have to create what we call an anonymous function.
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 5.848322 3.057333 3.758000 1.199333The general format of an anonymous function is function(x) body of the function.
For example, if you want to compute \(4^2\) using an anonymous function, it would be :
## [1] 16The anonymous function is surrounded by round brackets, and so is the input of the anonymous function.
Note : There is a shorter way to write anonymous functions :
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 5.848322 3.057333 3.758000 1.199333The function(df) is replaced by ~ and the argument of the function is replaced by a ..
17.5.4 Mapping with two data objects
So far, we have only used the purrr:map function that only takes one data object and one function as an argument.
What if we wanted to do more complicated operations, that use a function that needs more than one input?
For example, how would you calculate the weighted means (using weighted.mean) of the columns of a given data frame, where the weights are in another data frame?
Let’s create a data frame that contains the weights picking some randomly generated values from the iris_NA dataset (according to the poisson distribution using the rpois function) :
weights <- tibble(weight_sepal_legth = rpois(nrow(iris_NA), 3),
weight_sepal_width = rpois(nrow(iris_NA), 3),
weight_petal_legth = rpois(nrow(iris_NA), 3),
weight_petal_width = rpois(nrow(iris_NA), 3),)First, let’s see what are the parameters of weighted.mean
In order to know which purrr:map* function we have to use, you can consult the handy table where each row is the table corresponds to “the thing you want to map”.
Each column represents the type you want the “output” of the map function to be, either a list, an atomic (vector), the same type as the input, and no output (useful if you want to modify things in place).

Source: Advanced R by Hadley Wickham.
As we have two arguments, we should use the purrr:map2* function. As we want the output of the function to be a data frame, we are goint to use purrr:map2_df.
## # A tibble: 1 x 4
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## <dbl> <dbl> <dbl> <dbl>
## 1 NA 3.05 3.70 1.21We have the same issue as before because of the NAs… We should use an anonymous function!
## # A tibble: 1 x 4
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## <dbl> <dbl> <dbl> <dbl>
## 1 5.84 3.05 3.70 1.21What would be the short form of this anonymous function?
## # A tibble: 1 x 4
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## <dbl> <dbl> <dbl> <dbl>
## 1 5.84 3.05 3.70 1.21WARNING : if y has less elements than x, the elements of y will be used several times.
This could have some nasty side-effects, but is also quite useful!
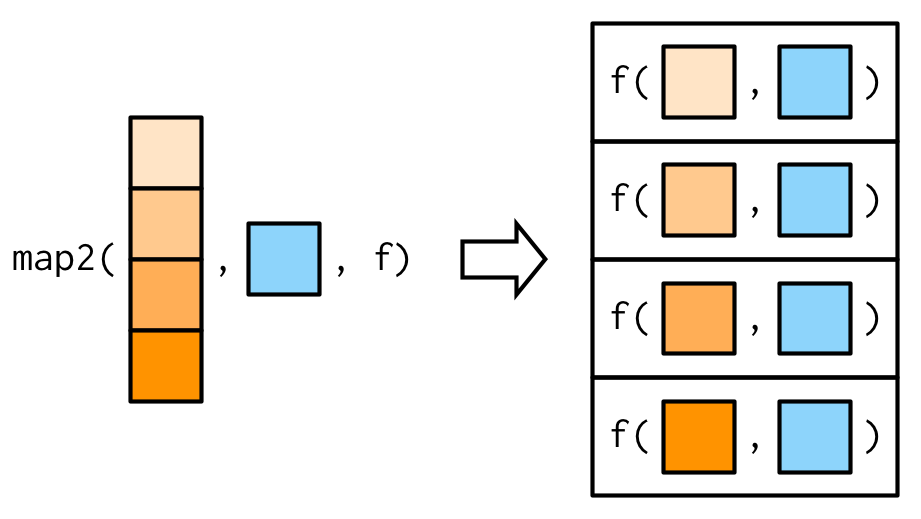 Source: Advanced R by Hadley Wickham.
Source: Advanced R by Hadley Wickham.
17.5.5 Mapping with more than two data objects
When we have more than two arguments, we should use the purrr:pmap* function.
## [[1]]
## [1] 1
##
## [[2]]
## [1] 2If we want to use an anonymous function, we have to us ..1, ..2, ..3.
## [[1]]
## [1] 3
##
## [[2]]
## [1] 4Note : if you use purrr:pmap* on a single data frame, it will iterate row-wise!
Example : Try to find the mean of all the rows of the iris_df dataset (which doesn’t really make sense, but let’s do it anyway).
## [[1]]
## [1] 5.1
##
## [[2]]
## [1] 4.9
##
## [[3]]
## [1] 4.7
##
## [[4]]
## [1] 4.6
##
## [[5]]
## [1] 5
##
## [[6]]
## [1] 5.417.5.6 Summary and key points
- Cupcakes (vanilla and chocolate and espresso) as motivation for writing functions in R
- The anatomy of an R function
- Vectorize - Which R functions are vectorized? What vectorization means.
- The benefits of vectorization (100M size vector takes 32s in a for loop, and <1 s in a vectorized function)
- Use the
purrrpackage to “map” over dataframes, vectors, etc… - There were more than 1 map_* ; choose the one that’s appropriate for your use case
- map2 and pmap! - as well as how to write anonymous functions in R
17.5.7 Additional Resources
- Chapter 21 of R for Data Science.
- Learn to purr blog post.
- Chapter 9 of Advanced R for Data Science.