Class Meeting 7 Intro to data wrangling, Part II
suppressPackageStartupMessages(library(tidyverse))
suppressPackageStartupMessages(library(gapminder))
suppressPackageStartupMessages(library(lubridate))
suppressPackageStartupMessages(library(tsibble))
suppressPackageStartupMessages(library(here))
suppressPackageStartupMessages(library(DT))7.1 Orientation (5 min)
7.1.1 Worksheet
You can find a worksheet template for today here.
7.1.2 Announcements
- Due tonight:
- Peer review 1 due tonight.
- Homework 2 due tonight.
- Reminder about setting up your own hw repo.
- stat545.com
7.1.3 Follow-up
- By the way, STAT 545 jumps into the tidyverse way of doing things in R, instead of the base R way of doing things. Lecture 2 was about “just enough” base R to get you started. If you feel that you want more practice here, take a look at the R intro stat videos by MarinStatsLectures (link is now in cm002 notes too).
ifstatement on its own won’t work withinmutate()because it’s not vectorized. Would need to vectorize it withsapply()or, better, thepurrr::mapfamily:
tibble(a = 1:4) %>%
mutate(b = (if (a < 3) "small" else "big"),
c = sapply(a, function(x) if (x < 3) "small" else "big"),
d = purrr::map_chr(a, ~ (if(.x < 3) "small" else "big")))## Warning in if (a < 3) "small" else "big": the condition has length > 1 and only
## the first element will be used## # A tibble: 4 x 4
## a b c d
## <int> <chr> <chr> <chr>
## 1 1 small small small
## 2 2 small small small
## 3 3 small big big
## 4 4 small big big7.1.4 Today’s Lessons
Where we are with dplyr:
Today: the unchecked boxes.
We’ll then look to a special type of tibble called a tsibble, useful for handling data with a time component. In doing so, we will touch on its older cousin, lubridate, which makes handling dates and times easier.
7.1.5 Resources
Concepts from today’s class are closely mirrored by the following resources.
- Jenny’s tutorial on Single table dplyr functions
Other resources:
- Like learning from a textbook? Check out all of r4ds: transform.
- The intro to
dplyrvignette is also a great resource.
Resources for specific concepts:
- To learn more about window functions and how dplyr handles them, see the window-functions vignette for the
dplyrpackage.
7.2 summarize() (3 min)
Like mutate(), the summarize() function also creates new columns, but the calculations that make the new columns must reduce down to a single number.
For example, let’s compute the mean and standard deviation of life expectancy in the gapminder data set:
## # A tibble: 1 x 2
## mu sigma
## <dbl> <dbl>
## 1 59.5 12.9Notice that all other columns were dropped. This is necessary, because there’s no obvious way to compress the other columns down to a single row. This is unlike mutate(), which keeps all columns, and more like transmute(), which drops all other columns.
As it is, this is hardly useful. But that’s outside of the context of grouping, coming up next.
7.3 group_by() (20 min)
The true power of dplyr lies in its ability to group a tibble, with the group_by() function. As usual, this function takes in a tibble and returns a (grouped) tibble.
Let’s group the gapminder dataset by continent and year:
## # A tibble: 1,704 x 6
## # Groups: continent, year [60]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # … with 1,694 more rowsThe only thing different from a regular tibble is the indication of grouping variables above the tibble. This means that the tibble is recognized as having “chunks” defined by unique combinations of continent and year:
- Asia in 1952 is one chunk.
- Asia in 1957 is another chunk.
- Europe in 1952 is another chunk.
- etc…
Notice that the data frame isn’t rearranged by chunk!
Now that the tibble is grouped, operations that you do on a grouped tibble will be done independently within each chunk, as if no other chunks exist.
You can also create new variables and group by that variable simultaneously. Try splitting life expectancy by “small” and “large” using 60 as a threshold:
## # A tibble: 1,704 x 7
## # Groups: smallLifeExp [2]
## country continent year lifeExp pop gdpPercap smallLifeExp
## <fct> <fct> <int> <dbl> <int> <dbl> <lgl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. TRUE
## 2 Afghanistan Asia 1957 30.3 9240934 821. TRUE
## 3 Afghanistan Asia 1962 32.0 10267083 853. TRUE
## 4 Afghanistan Asia 1967 34.0 11537966 836. TRUE
## 5 Afghanistan Asia 1972 36.1 13079460 740. TRUE
## 6 Afghanistan Asia 1977 38.4 14880372 786. TRUE
## 7 Afghanistan Asia 1982 39.9 12881816 978. TRUE
## 8 Afghanistan Asia 1987 40.8 13867957 852. TRUE
## 9 Afghanistan Asia 1992 41.7 16317921 649. TRUE
## 10 Afghanistan Asia 1997 41.8 22227415 635. TRUE
## # … with 1,694 more rows7.3.1 Grouped summarize() (10 min)
Want to compute the mean and standard deviation for each year for every continent? No problem:
## # A tibble: 60 x 4
## # Groups: continent [5]
## continent year mu sigma
## <fct> <int> <dbl> <dbl>
## 1 Africa 1952 39.1 5.15
## 2 Africa 1957 41.3 5.62
## 3 Africa 1962 43.3 5.88
## 4 Africa 1967 45.3 6.08
## 5 Africa 1972 47.5 6.42
## 6 Africa 1977 49.6 6.81
## 7 Africa 1982 51.6 7.38
## 8 Africa 1987 53.3 7.86
## 9 Africa 1992 53.6 9.46
## 10 Africa 1997 53.6 9.10
## # … with 50 more rowsNotice:
- The grouping variables are kept in the tibble, because their values are unique within in chunk (by definition of the chunk!)
- With each call to
summarize(), the grouping variables are “peeled back” from last grouping variable to first.
This means the above tibble is now only grouped by continent. What happens when we reverse the grouping?
gapminder %>%
group_by(year, continent) %>% # Different order
summarize(mu = mean(lifeExp),
sigma = sd(lifeExp))## # A tibble: 60 x 4
## # Groups: year [12]
## year continent mu sigma
## <int> <fct> <dbl> <dbl>
## 1 1952 Africa 39.1 5.15
## 2 1952 Americas 53.3 9.33
## 3 1952 Asia 46.3 9.29
## 4 1952 Europe 64.4 6.36
## 5 1952 Oceania 69.3 0.191
## 6 1957 Africa 41.3 5.62
## 7 1957 Americas 56.0 9.03
## 8 1957 Asia 49.3 9.64
## 9 1957 Europe 66.7 5.30
## 10 1957 Oceania 70.3 0.0495
## # … with 50 more rowsThe grouping columns are switched, and now the tibble is grouped by year instead of continent.
dplyr has a bunch of convenience functions that help us write code more eloquently. We could use group_by() and summarize() with length() to find the number of entries each country has:
## # A tibble: 1,704 x 2
## # Groups: country [142]
## country n
## <fct> <int>
## 1 Afghanistan 12
## 2 Afghanistan 12
## 3 Afghanistan 12
## 4 Afghanistan 12
## 5 Afghanistan 12
## 6 Afghanistan 12
## 7 Afghanistan 12
## 8 Afghanistan 12
## 9 Afghanistan 12
## 10 Afghanistan 12
## # … with 1,694 more rowsOr, we can use dplyr::n() to count the number of rows in each group:
## # A tibble: 142 x 2
## country n
## <fct> <int>
## 1 Afghanistan 12
## 2 Albania 12
## 3 Algeria 12
## 4 Angola 12
## 5 Argentina 12
## 6 Australia 12
## 7 Austria 12
## 8 Bahrain 12
## 9 Bangladesh 12
## 10 Belgium 12
## # … with 132 more rowsOr better yet, just use dplyr::count():
## # A tibble: 142 x 2
## country n
## <fct> <int>
## 1 Afghanistan 12
## 2 Albania 12
## 3 Algeria 12
## 4 Angola 12
## 5 Argentina 12
## 6 Australia 12
## 7 Austria 12
## 8 Bahrain 12
## 9 Bangladesh 12
## 10 Belgium 12
## # … with 132 more rows7.3.2 Grouped mutate() (3 min)
Want to get the increase in GDP per capita for each country? No problem:
gap_inc <- gapminder %>%
arrange(year) %>%
group_by(country) %>%
mutate(gdpPercap_inc = gdpPercap - lag(gdpPercap))
DT::datatable(gap_inc)You can’t see it here (because of the datatable() output), but the tibble is still grouped by country.
Drop the NAs with another convenience function, this time supplied by the tidyr package (another tidyverse package that we’ll see soon):
## # A tibble: 1,562 x 7
## # Groups: country [142]
## country continent year lifeExp pop gdpPercap gdpPercap_inc
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Afghanistan Asia 1957 30.3 9240934 821. 41.4
## 2 Albania Europe 1957 59.3 1476505 1942. 341.
## 3 Algeria Africa 1957 45.7 10270856 3014. 565.
## 4 Angola Africa 1957 32.0 4561361 3828. 307.
## 5 Argentina Americas 1957 64.4 19610538 6857. 946.
## 6 Australia Oceania 1957 70.3 9712569 10950. 910.
## 7 Austria Europe 1957 67.5 6965860 8843. 2706.
## 8 Bahrain Asia 1957 53.8 138655 11636. 1769.
## 9 Bangladesh Asia 1957 39.3 51365468 662. -22.6
## 10 Belgium Europe 1957 69.2 8989111 9715. 1372.
## # … with 1,552 more rows7.4 Function types (5 min)
We’ve seen cases of transforming variables using mutate() and summarize(), both with and without group_by(). How can you know what combination to use? Here’s a summary based on one of three types of functions.
| Function type | Explanation | Examples | In dplyr |
|---|---|---|---|
| Vectorized functions | These take a vector, and operate on each component independently to return a vector of the same length. In other words, they work element-wise. | cos(), sin(), log(), exp(), round() |
mutate() |
| Aggregate functions | These take a vector, and return a vector of length 1 | mean(), sd(), length() |
summarize(), esp with group_by(). |
| Window Functions | these take a vector, and return a vector of the same length that depends on the vector as a whole. | lag(), rank(), cumsum() |
mutate(), esp with group_by() |
7.5 dplyr Exercises (20 min)
7.6 Dates and Times (5 min)
The lubridate package is great for identifying dates and times. You can also do arithmetic with dates and times with the package, but we won’t be discussing that.
Make an object of class "Date" using a function that’s some permutation of y, m, and d (for year, month, and date, respectively). These functions are more flexible than your yoga instructor:
## [1] "2019-09-24"## [1] "2019-09-24"## [1] "2019-09-24"## [1] "2019-09-24" "2019-09-25"Notice that they display the dates all in ymd format, which is best for computing because the dates sort nicely this way.
This is not just a character!
## [1] "Date"You can tag on hms, too:
## [1] "2019-09-24 23:59:59 UTC"We can also extract information from these objects. Day of the week:
## [1] Tue
## Levels: Sun < Mon < Tue < Wed < Thu < Fri < SatDay:
## [1] 24Number of days into the year:
## [1] 267Is it a leap year this year?
## [1] FALSEThe newer tsibble package gives these lubridate functions some friends. What’s the year and month? Year and week?
## [1] "2019 Sep"## [1] "2019 W39"7.7 Tsibbles (15 min)
A tsibble (from the package of the same name) is a special type of tibble, useful for handling data where a column indicates a time variable.
As an example, here are daily records of a household’s electricity usage:
## Parsed with column specification:
## cols(
## date = col_date(format = ""),
## intensity = col_double(),
## voltage = col_double()
## )## # A tibble: 1,360 x 3
## date intensity voltage
## <date> <dbl> <dbl>
## 1 2007-01-01 14.6 246.
## 2 2008-01-01 24.2 247.
## 3 2009-01-01 36.4 250.
## 4 2010-01-01 20.6 249.
## 5 2007-10-01 33.2 246.
## 6 2008-10-01 26.2 248.
## 7 2009-10-01 22.8 248.
## 8 2010-10-01 18.2 247.
## 9 2007-11-01 6 246.
## 10 2008-11-01 7 247.
## # … with 1,350 more rowsLet’s make this a tsibble in the same way we’d convert a data frame to a tibble: with the as_tsibble() function. The conversion requires you to specify which column contains the time index, using the index argument.
## # A tsibble: 1,360 x 3 [1D]
## date intensity voltage
## <date> <dbl> <dbl>
## 1 2006-12-16 33.2 244.
## 2 2006-12-17 30 249.
## 3 2006-12-18 27 248.
## 4 2006-12-19 33.6 249.
## 5 2006-12-20 25.2 249.
## 6 2006-12-22 34.2 249.
## 7 2006-12-23 37 247.
## 8 2006-12-24 29.6 249.
## 9 2006-12-25 28.4 251.
## 10 2006-12-26 27.4 249.
## # … with 1,350 more rowsWe already see an improvement vis-a-vis the sorted dates!
This is an example of time series data, because the time interval has a regular spacing. A tsibble cleverly determines and stores this interval. With the energy consumption data, the interval is one day (“1D” means “1 day”, not “1 dimension”!):
## 1DNotice that there is no record for December 21, 2006, in what would be Row 5. Such records are called implicit NA’s, because they’re actually missing, but aren’t explicitly shown as missing in your data set. If you don’t make these explicit, you could mess up your analysis if it’s anticipating your data to be equally spaced in time. Just full_gaps() to bring them out of hiding:
## # A tsibble: 1,442 x 3 [1D]
## date intensity voltage
## <date> <dbl> <dbl>
## 1 2006-12-16 33.2 244.
## 2 2006-12-17 30 249.
## 3 2006-12-18 27 248.
## 4 2006-12-19 33.6 249.
## 5 2006-12-20 25.2 249.
## 6 2006-12-21 NA NA
## 7 2006-12-22 34.2 249.
## 8 2006-12-23 37 247.
## 9 2006-12-24 29.6 249.
## 10 2006-12-25 28.4 251.
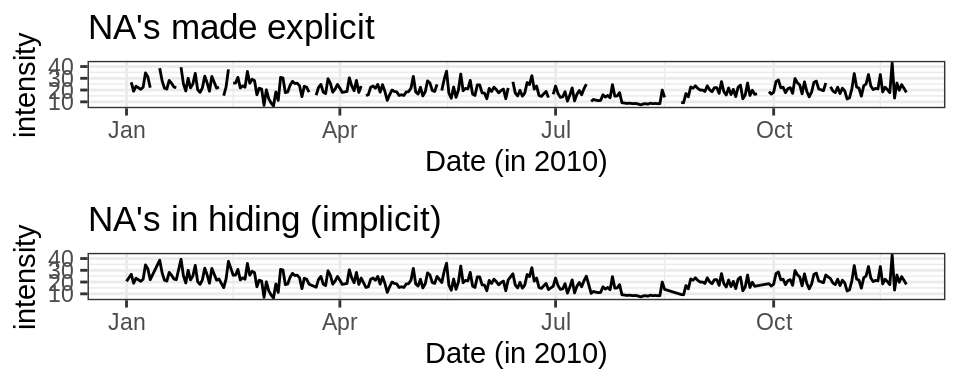
## # … with 1,432 more rowsAlready, it’s better to plot the data now that these gaps are filled in. Let’s check out 2010. See how the plot without NA’s can be a little misleading? Moral: always be as honest as possible with your data.
small_energy <- filter(energy, year(date) == 2010)
cowplot::plot_grid(
ggplot(small_energy, aes(date, intensity)) +
geom_line() +
theme_bw() +
xlab("Date (in 2010)") +
ggtitle("NA's made explicit"),
ggplot(drop_na(small_energy), aes(date, intensity)) +
geom_line() +
theme_bw() +
xlab("Date (in 2010)") +
ggtitle("NA's in hiding (implicit)"),
nrow = 2
)
How would we convert gapminder to a tsibble, since it has a time series per country? Use the key argument to specify the grouping:
## # A tsibble: 1,704 x 6 [5Y]
## # Key: country [142]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # … with 1,694 more rows7.7.1 index_by() instead of group_by() (5 min)
It looks like there’s seasonality in intensity across the year:
ggplot(energy, aes(yday(date), intensity)) +
geom_point() +
theme_bw() +
labs(x = "Day of the Year")## Warning: Removed 82 rows containing missing values (geom_point).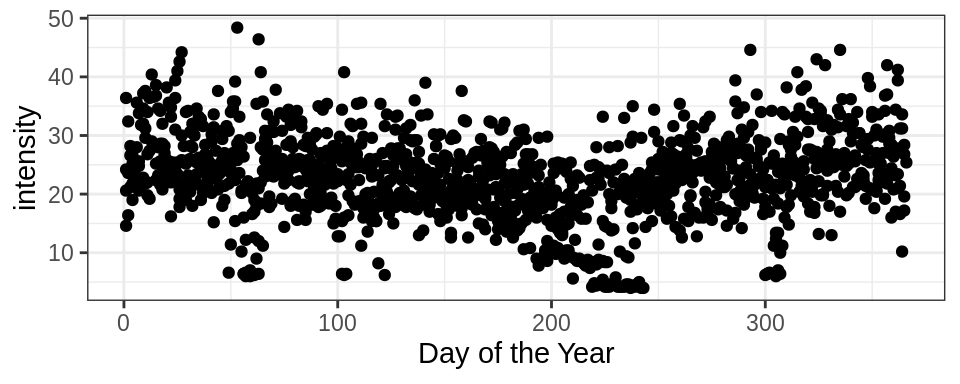
Let’s get a mean estimate of intensity on each day of the year. We’d like to group_by(yday(date)), but because we’re grouping on the index variable, we use index_by() instead.
energy %>%
tsibble::index_by(day_of_year = yday(date)) %>%
dplyr::summarize(mean_intensity = mean(intensity, na.rm = TRUE))## # A tsibble: 366 x 2 [1]
## day_of_year mean_intensity
## <dbl> <dbl>
## 1 1 24.0
## 2 2 24.1
## 3 3 25.0
## 4 4 23.2
## 5 5 23.4
## 6 6 27.6
## 7 7 26.2
## 8 8 24.4
## 9 9 31.6
## 10 10 30.0
## # … with 356 more rowsWhat if we wanted to make the time series less granular? Instead of total daily consumption, how about total weekly consumption? Note the convenience function summarize_all() given to us by dplyr!
## # A tsibble: 207 x 3 [1W]
## yearweek intensity voltage
## <week> <dbl> <dbl>
## 1 2006 W50 63.2 493.
## 2 2006 W51 NA NA
## 3 2006 W52 NA NA
## 4 2007 W01 172. 1737.
## 5 2007 W02 NA NA
## 6 2007 W03 211. 1731.
## 7 2007 W04 NA NA
## 8 2007 W05 197 1731.
## 9 2007 W06 183. 1732.
## 10 2007 W07 194. 1732.
## # … with 197 more rowsBy the way, there’s no need to worry about “truncated weeks” at the beginning and end of the year. For example, December 31, 2019 is a Tuesday, and is Week 53, but its “yearmonth” is Week 1 in 2020:
## [1] Tue
## Levels: Sun < Mon < Tue < Wed < Thu < Fri < Sat## [1] 53## [1] "2020 W01"